SLIM MICROSCOPY ALLOWS FOR VISUALIZATION OF DNA-CONTAINING LIPOSOMES DESIGNED FOR SPERM-MEDIATED GENE TRANSFER IN CATTLE
Marcello Rubessa, Samantha N. Lotti, Mikhail E. Kandel, Gabriel Popescu, Mathew B. Wheeler
Molecular Biology Reports 2018 695-703
![]()
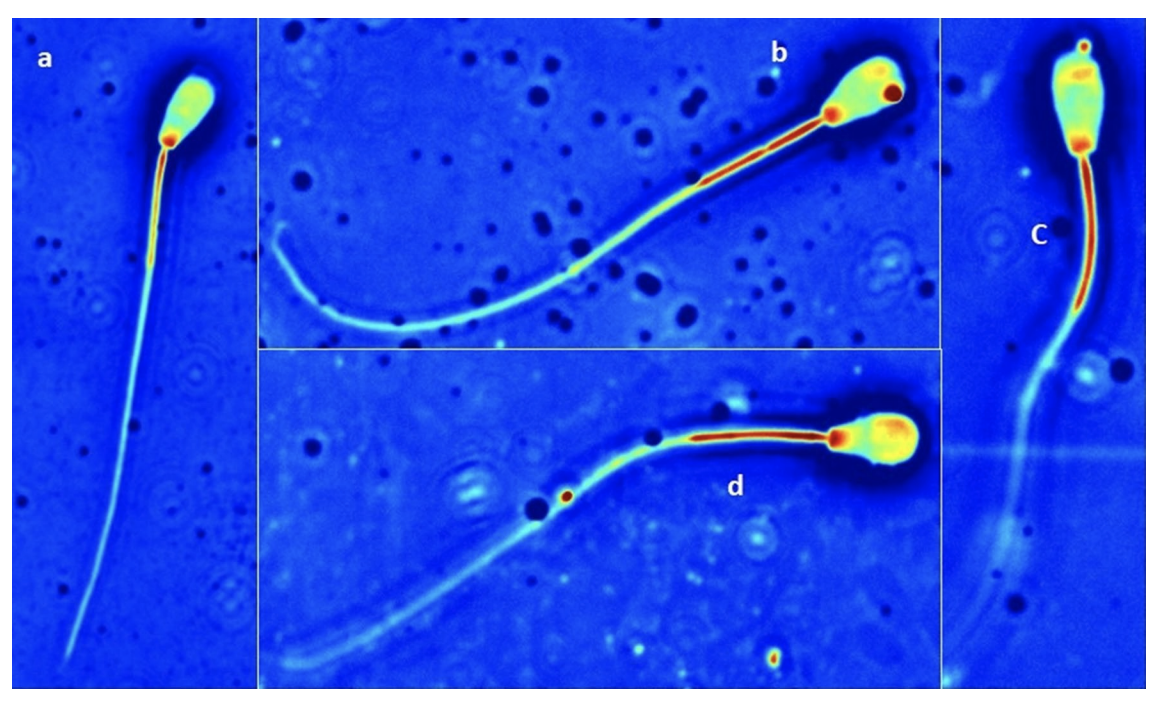
Naked DNA has been shown to bind naturally to the sperm, a method called sperm-mediated gene transfer (SMGT). Based on these observations, we examined the efficiency of exogenous DNA binding to sperm using liposomes. In this experiment, we analyzed methods to select frozen-thawed bovine sperm, and evaluated the binding of exogenous DNA to those sperm. To determine the optimal selection method, we used Computer-Assisted Sperm Analysis (CASA). Percoll or Swim-Up were used to select sperm, followed by incubation up to 3 h with the liposome-DNA complexes. The samples were collected after 1 h and after 3 h. We used enhanced green fluorescent protein (eGFP) in combination with the liposomes as a marker for exogenous DNA binding. Five treatments per selection method were analyzed: (1) no incubation, no liposomes and no DNA, (2) incubation with no liposomes and no DNA, (3) incubation with liposomes and no DNA, (4) incubation with liposomes and 1 µg of DNA and (5) incubation with liposomes and 10 µg of DNA. The CASA results for total motility and rapid motility were statistically significant (P < 0.01) between the control and the other treatments in the Percoll group as opposed to Swim-Up. Swim-Up was therefore chosen as the optimal selection method. In order to determine if the liposome-DNA complex had bound to sperm, real time PCR was used to detect GFP DNA and images of the sperm were analyzed using the Spatial Light Interference Microscopy (SLIM). SLIM confirmed the presence of liposomes on the sperm head and tail.
