Neuroscience using CellVista SLIM — Label-free proliferation assay and cell growth
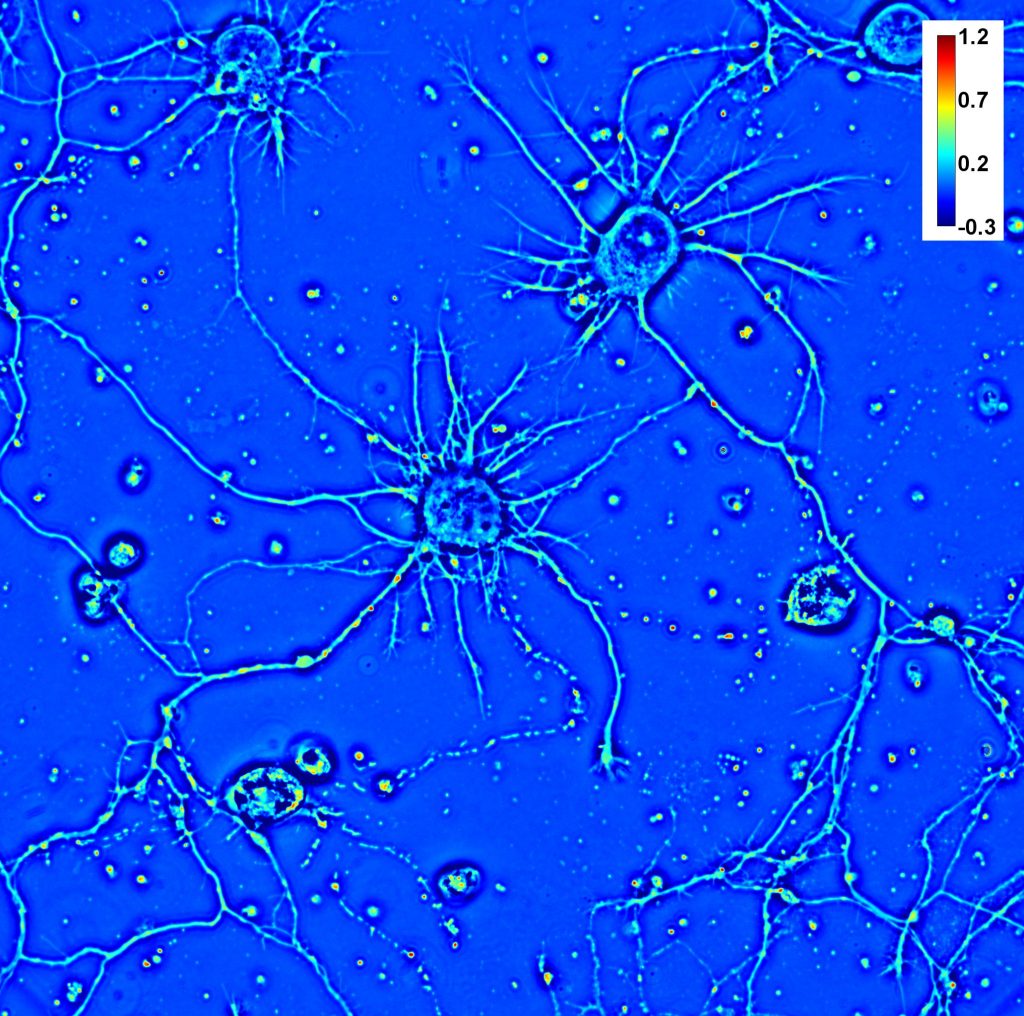
This Application highlights the use of CellVista SLIM (Spatial Light Interference Microscopy) for neuroscience research. The non-invasive live cell imaging provides a viable environment for fragile neurons and neuronal stem cells, which are very susceptible to damages from temperature, chemicals, and light.
The speed of CellVista SLIM acquisition is capable of detecting the transport between neurons, and the wide field of view is capable of imaging the formation of a neuronal network.
Therefore, CellVista SLIM provides an environment where neuronal cultures can be studied at both single cell level and population level.
We can produce live demonstrations for this topic, either with in-house samples or delivered samples.
Phi Optics CellVista System seamlessly upgrades the camera port of your microscope for neuroscience studies.
Free Pre-Recorded Webinar
Register to access this free online video webinar which covers this topic in-depth.
for Neuroscience


CellVista SLIM is a non-invasive phase imaging technology that quantifies the physical properties of live cells and tissues. The output is a live quantitative image (SLIM map) of the specimen on the microscope stage. The intensity of every pixel in the frame is a measure of the optical path length difference (in radians) through the sample, i.e. a phase shift map, which is measured with better than 0.5 nanometers sensitivity [1]. The phase shift map is converted on-the-fly to other SLIM maps, with their respective pixel intensity: thickness (in microns), dry mass area density (picograms per square micron) and refractive index. More detailed description of SLIM can be found in the reference pages in the How It Works page of this website.
.
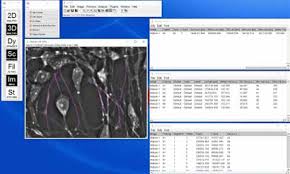
Figure 2. Setup of NeuronJ tracking in CellVista SLIM images from ImageJ
Simple measurements – NeuronJ
NeuronJ is an ImageJ plugin developed by Meijering et al. [2]. The plugin can be found in the 2D analysis menu, and once run, a new toolset on the main ImageJ window loads. NeuronJ allows the user to easily trace along neurites and performs measurements on neuronal cultures for parameters such as the length, mean value along the trace, and many more. Moreover, the traces can be saved as text file with the coordinates and values of the points along the trace, and allows the user to treat the data as a list of values instead of a complex structure. The steps to perform the measurements are as follows:
- Convert the SLIM map to an 8-bit image and save
- Run NeuronJ found in 2D analysis menu, load the 8-bit image using NeuronJ “Load images”
- Trace neurites using “Add tracings” in NeuronJ
- Run measurements using “Measure tracings” in NeuronJ (Figure 2)
Traffic
Rapid exchanges of material among cells in neuronal cultures appear in the form of vesicle transport along the neurites that connect neighboring neurons. These vesicles are higher in density compared to the neurites and appears as a particle moving along the neurites. Therefore, by applying a particle tracking method, the movement of these vesicles can be tracked and the transport or “traffic” along the neuronal highway can be studied [3]. For more details see our Cell Dynamics application page.
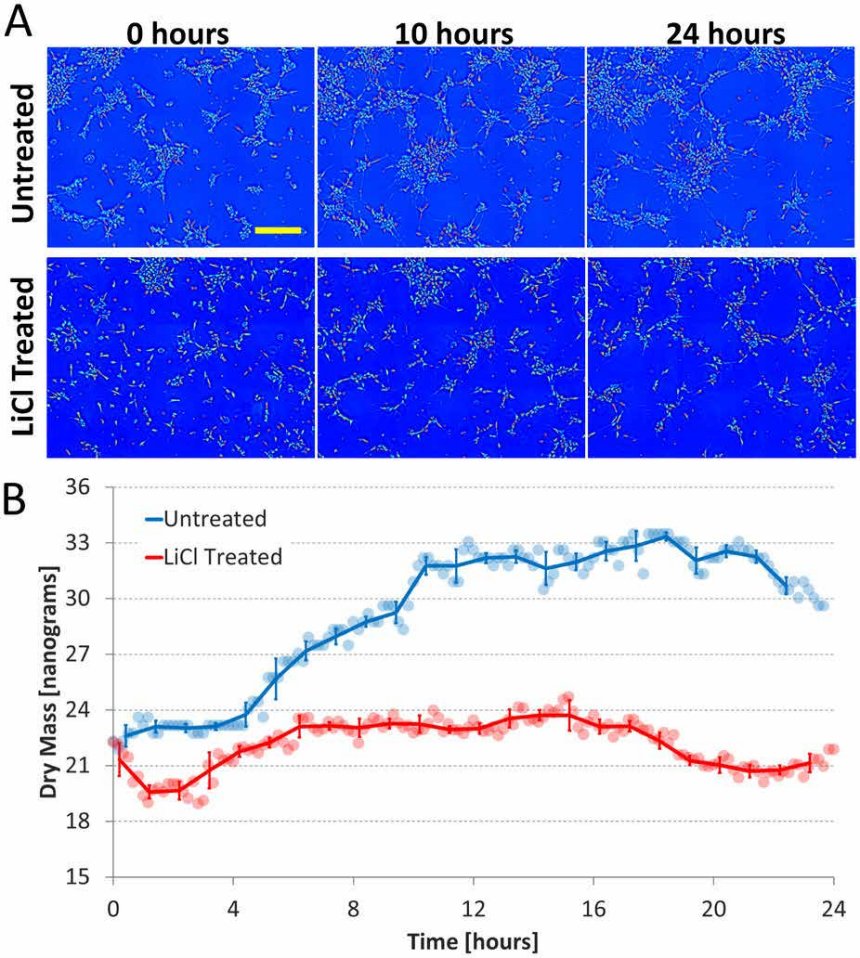
Figure 3. CellVista SLIM measures dry mass over time.
Growth
Because neuronal cultures are very fragile and vulnerable to many invasive factors, such as chemicals, light, and heat, it is difficult to study them live and study their growth.
CellVista SLIM provides a solution for this type of problem by providing an imaging environment that is both temperature and CO2 controlled, and also by exposing the cells to minimal light damage. The growth of neurons have been studied and the emergence of neuronal network have been shown using SLIM [4].
Figure on Left: Measuring cell growth over time and the effect of a growth inhibitor, lithium chloride (LiCl) using CellVista SLIM. (a) Dry mass density maps acquired at 0, 10, and 24 hours for both the untreated and LiCl treated cultures; the yellow scale bar corresponds to 200 um. (b) Total dry mass vs. time of the entire field of view for both conditions.
Round markers are raw mass data, solid lines are the average over 1 hour, and error bars indicate standard deviation. For the untreated culture it can be seen that majority of the mass growth occurs between 0 and 10 hours, the time during which the cells are extending processes most actively. In the LiCl treated culture neurite outgrowth is severely retarded and no significant increase in mass is observed during any period.
Differentiation
The non-invasive environment that CellVista SLIM provides is suitable for studying not only mature neuron cells but also progenitor cells, which are even more fragile than mature neurons. The structural change of the neural progenitor cells and the change in dynamics can easily be studied using CellVista SLIM to show the development of mature cells from the progenitor cells, while keeping the cell alive and well.
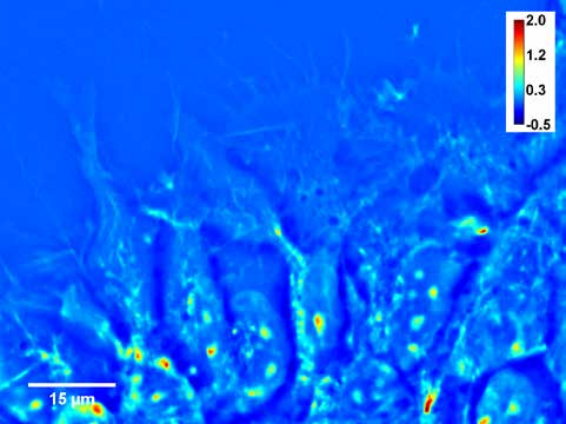
Figure. Neural progenitor cells plated, day 0
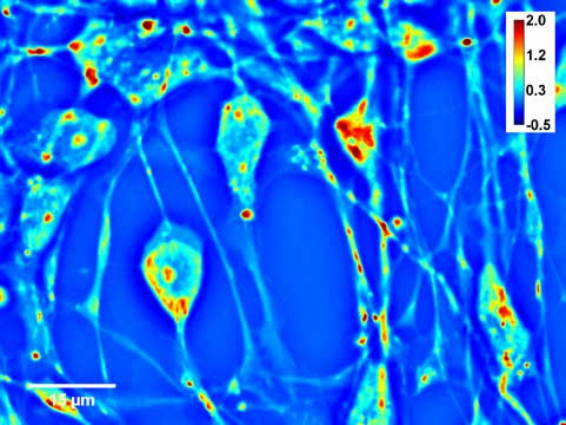
Figure. Neural progenitor cells plated, day 14
For high image resolution solutions of thick specimens, please see our CellVista GLIM™ systems. For customization don’t hesitate to CONTACT US!
References:
[1] G. Popescu (2011) Quantitative phase imaging of cells and tissues (McGrow-Hill, New York)
[2] E. Meijering, M. Jacob, J.-C. F. Sarria, P. Steiner, H. Hirling and M. Unser, Design and validation tool for neurite tracing and analysis in uorescence microscopy images, Cytometry Part A, 58 (2), 167-176 (2004).
[3] Z. Wang, L. Millet, V. Chan, H. Ding, M. U. Gillette, R. Bashir and G. Popescu, Label-free intracellular trans- port measured by spatial light interference microscopy, Journal of Biomedical Optics, 16 (2), 026019 (2011).
[4] M. Mir, T. Kim, A. Majumder, M. Xiang, R. Wang, S. C. Liu, M. U. Gillette, S. Stice and G. Popescu, Label-free characterization of emerging human neuronal networks, Scientific Reports, 4, 4434 (2014)
