REPRODUCTIVE OUTCOMES PREDICTED BY PHASE IMAGING WITH COMPUTATIONAL SPECIFICITY OF SPERMATOZOON ULTRASTRUCTURE
Mikhail E. Kandela,b,1, Marcello Rubessac,1, Yuchen R. Hea,b, Sierra Schreibera,c, Sasha Meyersa,c, Luciana Matter Navesc , Molly K. Sermersheimc , G. Scott Selld , Michael J. Szewczyka , Nahil Sobha , Matthew B. Wheelera,c,e,1, and Gabriel Popescua,b,e,1,2
PNAS, vol. 117, no. 31, 18309 2020
![]()
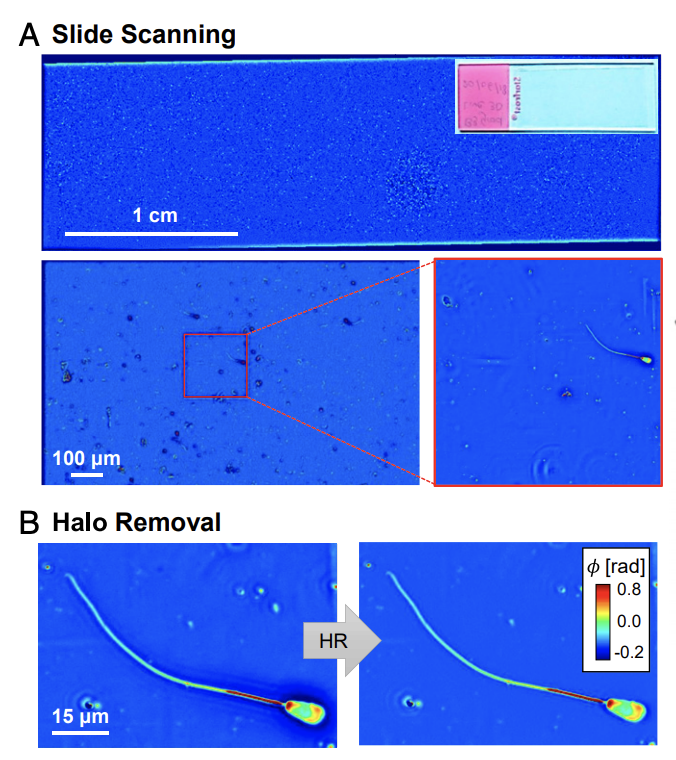
The ability to evaluate sperm at the microscopic level, at high-throughput, would be useful for assisted reproductive technologies (ARTs), as it can allow specific selection of sperm cells for in vitro fertilization (IVF). The tradeoff between intrinsic imaging and external contrast agents is particularly acute in reproductive medicine. The use of fluorescence labels has enabled new cell-sorting strategies and given new insights into developmental biology. Nevertheless, using extrinsic contrast agents is often too invasive for routine clinical operation. Raising questions about cell viability, especially for single-cell selection, clinicians prefer intrinsic contrast in the form of phase-contrast, differential-interference contrast, or Hoffman modulation contrast. While such instruments are nondestructive, the resulting image suffers from a lack of specificity. In this work, we provide a template to circumvent the tradeoff between cell viability and specificity by combining high-sensitivity phase imaging with deep learning. In order to introduce specificity to label-free images, we trained a deep-convolutional neural network to perform semantic segmentation on quantitative phase maps. This approach, a form of phase imaging with computational specificity (PICS), allowed us to efficiently analyze thousands of sperm cells and identify correlations between dry-mass content and artificial-reproduction outcomes. Specifically, we found that the dry-mass content ratios between the head, midpiece, and tail of the cells can predict the percentages of success for zygote cleavage and embryo blastocyst formation.

