LABEL-FREE, MULTI-SCALE IMAGING OF EX-VIVO MOUSE BRAIN USING SPATIAL LIGHT INTERFERENCE MICROSCOPY
Eunjung Min1,2, Mikhail E. Kandel1, CheMyong J Ko3, Gabriel Popescu1, Woonggyu Jung2,4 & Catherine Best-Popescu5
Scientific Reports, 6:39667 2016
![]()
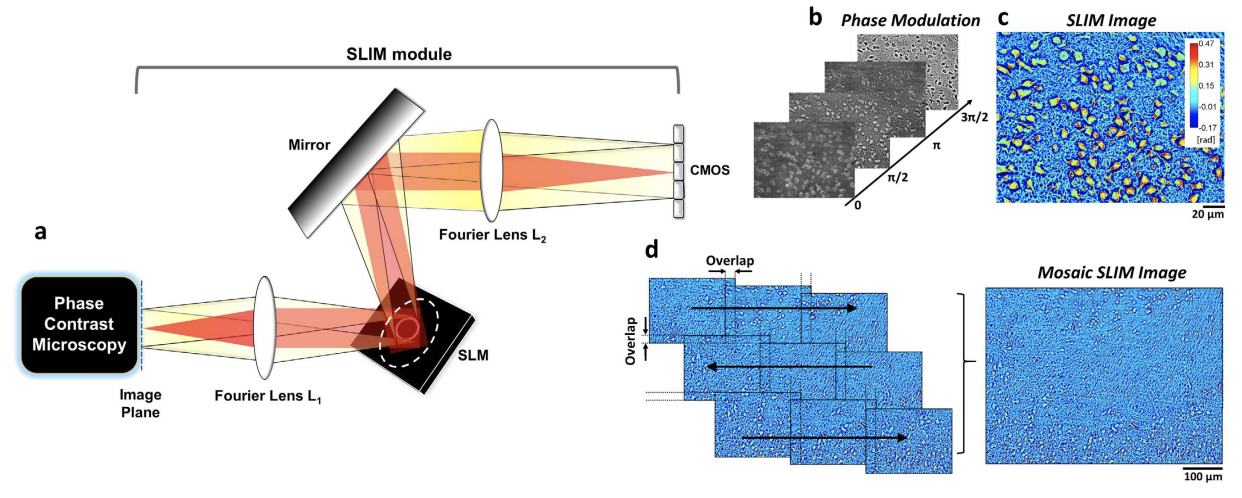
Brain connectivity spans over broad spatial scales, from nanometers to centimeters. In order to understand the brain at multi-scale, the neural network in wide-field has been visualized in detail by taking advantage of light microscopy. However, the process of staining or addition of fluorescent tags is commonly required, and the image contrast is insufficient for delineation of cytoarchitecture. To overcome this barrier, we use spatial light interference microscopy to investigate brain structure with high-resolution, sub-nanometer pathlength sensitivity without the use of exogenous contrast agents. Combining wide-field imaging and a mosaic algorithm developed in-house, we show the detailed architecture of cells and myelin, within coronal olfactory bulb and cortical sections, and from sagittal sections of the hippocampus and cerebellum. Our technique is well suited to identify laminar characteristics of fiber tract orientation within white matter, e.g. the corpus callosum. To further improve the macro-scale contrast of anatomical structures, and to better differentiate axons and dendrites from cell bodies, we mapped the tissue in terms of its scattering property. Based on our results, we anticipate that spatial light interference microscopy can potentially provide multiscale and multicontrast perspectives of gross and microscopic brain anatomy.

