LABEL-FREE COLORECTAL CANCER SCREENING USING DEEP LEARNING AND SPATIAL LIGHT INTERFERENCE MICROSCOPY (SLIM)
Jingfang “Kelly” Zhang1,3, Yuchen He1,2,3, Nahil Sobh1,3, and Gabriel Popescu1,2,3,4
APL Photonics, 10, 1063-5, 004723 2020
![]()
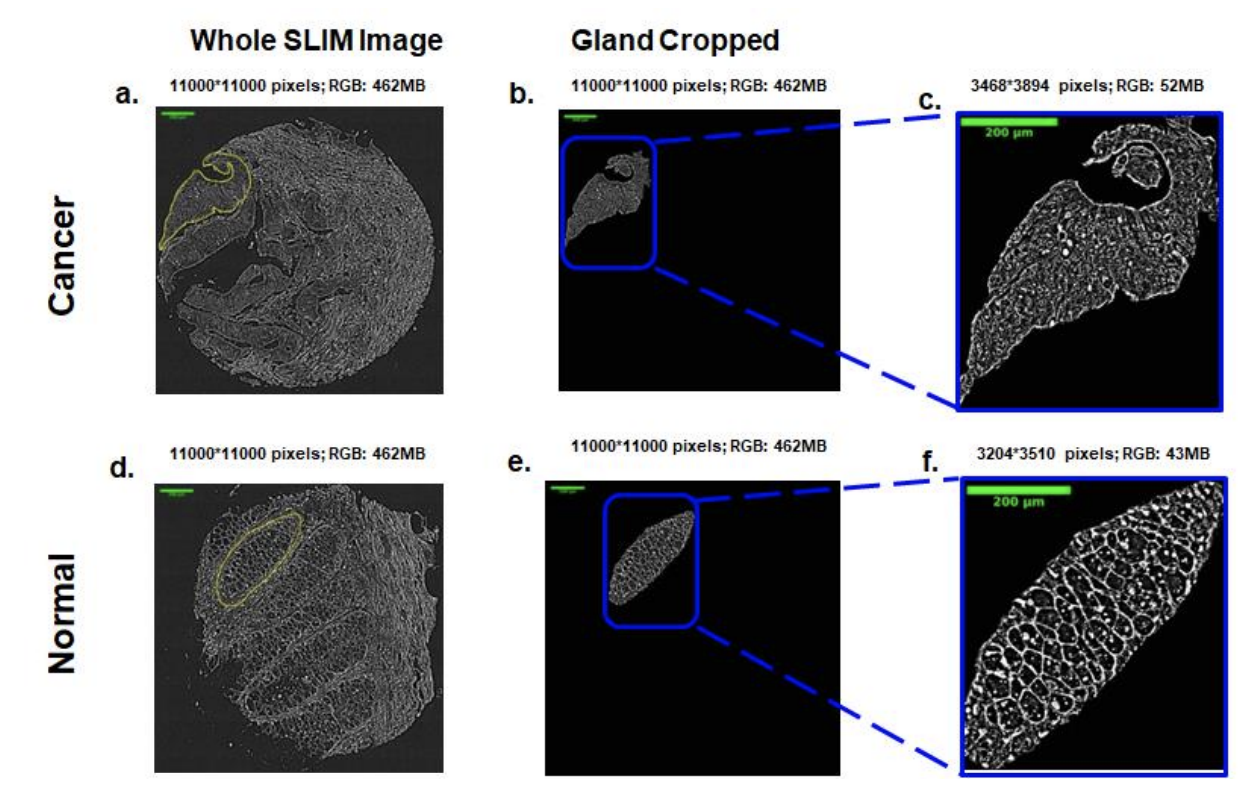
Current pathology workflow involves staining of thin tissue slices, which otherwise would be transparent, followed by manual investigation under the microscope by a trained pathologist. While the hematoxylin and eosin (H&E) stain is well-established and a cost-effective method for visualizing histology slides, its color variability across preparations and subjectivity across clinicians remain unaddressed challenges. To mitigate these challenges, recently, we have demonstrated that spatial light interference microscopy (SLIM) can provide a path to intrinsic objective markers that are independent of preparation and human bias. Additionally, the sensitivity of SLIM to collagen fibers yields information relevant to patient outcome, which is not available in H&E. Here, we show that deep learning and SLIM can form a powerful combination for screening applications: training on 1660 SLIM images of colon glands and validating on 144 glands, we obtained an accuracy of 98% (validation dataset) and 99% (test dataset), resulting in benign vs cancer classification accuracy of 97%, defined as area under the receiver operating characteristic curve. We envision that the SLIM whole slide scanner presented here paired with artificial intelligence algorithms may prove valuable as a pre-screening method, economizing the clinician’s time and effort.

