PHASE IMAGING WITH COMPUTATIONAL SPECIFICITY (PICS) FOR MEASURING DRY MASS CHANGES IN SUB-CELLULAR COMPARTMENTS
Mikhail E. Kandel 1,2,8, Yuchen R. He 1,2,8, Young Jae Lee1,3, Taylor Hsuan-Yu Chen1,4, Kathryn Michele Sullivan4, Onur Aydin5, M. Taher A. Saif4,5, Hyunjoon Kong1,4,6,7, Nahil Sobh 1✉ & Gabriel Popescu 1,2,4✉
Nature Comm. 11, Article 6256 2020
![]()
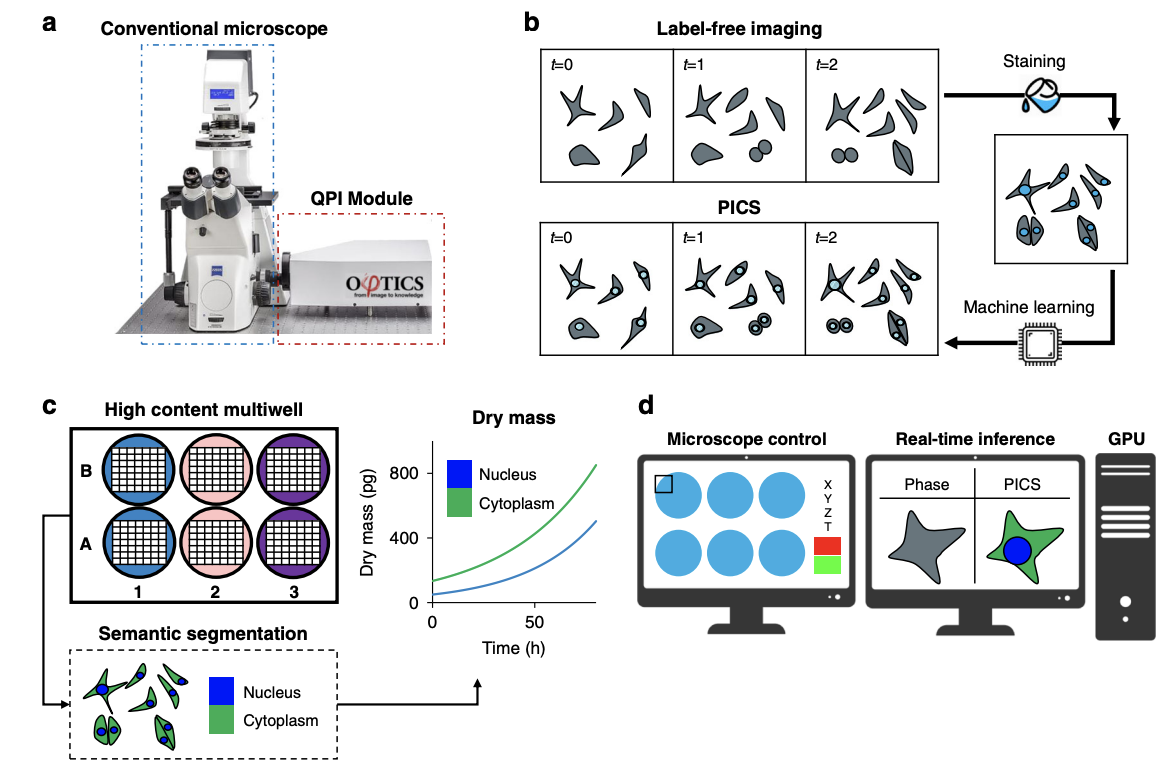
Due to its specificity, fluorescence microscopy has become a quintessential imaging tool in cell biology. However, photobleaching, phototoxicity, and related artifacts continue to limit fluorescence microscopy’s utility. Recently, it has been shown that artificial intelligence (AI) can transform one form of contrast into another. We present phase imaging with computational specificity (PICS), a combination of quantitative phase imaging and AI, which provides information about unlabeled live cells with high specificity. Our imaging system allows for automatic training, while inference is built into the acquisition software and runs in real-time. Applying the computed fluorescence maps back to the quantitative phase imaging (QPI) data, we measured the growth of both nuclei and cytoplasm independently, over many days, without loss of viability. Using a QPI method that suppresses multiple scattering, we measured the dry mass content of individual cell nuclei within spheroids. In its current implementation, PICS offers a versatile quantitative technique for continuous simultaneous monitoring of individual cellular components in biological applications where long-term label-free imaging is desirable.
